
SimePhys

 | SimePhys |  |
|
This is an application to learn and simulate basic electrophysiological experiments
of in vivo preparations of the blow fly's visual system. Current version is: V1.2.1 (stable) from 28-Dec-2025 V1.4.5 (experimental) from 31-Dec-2022 --
Table of ContentWelcome - a brief introduction about the program License - terms of use Requirements - what is needed to run this program? Installation - installation of the program Usage - how to use the programm *.fly viewer - a tool to monitor your recorded data FAQ - frequently asked questions Download - get your copy here
History - history and future develoment
This is an application to learn and simulate basic electrophysiological experiments of in vivo preparations. While the main purpose of this software is teaching and learning, it is also some kind of test ballon for neuronal coding and signaling models.
The software simulates experiments like performed on the visual system of the fly.
There are several well characterized
cells (currently H1 of both hemispheres, V1 and V2)
implemented inside the software that respond to moving grating patterns.
Moreover there are many not exactly specified cells implemented counting to a total of 45 cells.
Cell recordings are done extracellularly. For background information about the adaptation experiments please refer to T. Maddess and S.B. Laughlin (1985): Adaptation of Motion-Sensitive Neuron H1 is Generated Locally and Governed by Contrast Frequency. Proc. R. Soc. Lond. B. 225: 252-275 DOI:10.1098/rspb.1985.0061 For further information don't hesitate to contact me. I am happy for any feedback. This software currently is free for personal use. You may use it on your own risk. You are free to copy and distribute the archive. When distributing and/or spreading the application you must not:
This software is written in the Hollywood programming language and thus
available for many systems.
There is no additional
software required and it should run on bog standard computers running one of the
supported operating systems. You may need a processor better not less than 800 MHz
and maybe a few MB (roughly about 25, depends on the particular system) of available
RAM. To sum it up, if you don't use
something very special or antique you probably will be able to run this program. The operating systems supported are: Binaries for OS X, Linux, AmigaOS and AROS and are not provided on this site but are available upon request only. If you run the *.fly viewer without Simephys you need less processor power and a resolution of 1024x600 is sufficient (ideal for usual netbooks). Once you've downloaded the archive for your operating system, unpack the archive and copy it to any destination you like. You may put it on your harddsik or on a usb key or even on a server. There is no special installation process required
Just doubleclick the icon named Simephys to launch the program. Once the splash screen disappeared (usually quick, but can take a few seconds) the progam launches three windows as shown in the screen shots below (MorphOS version):
|
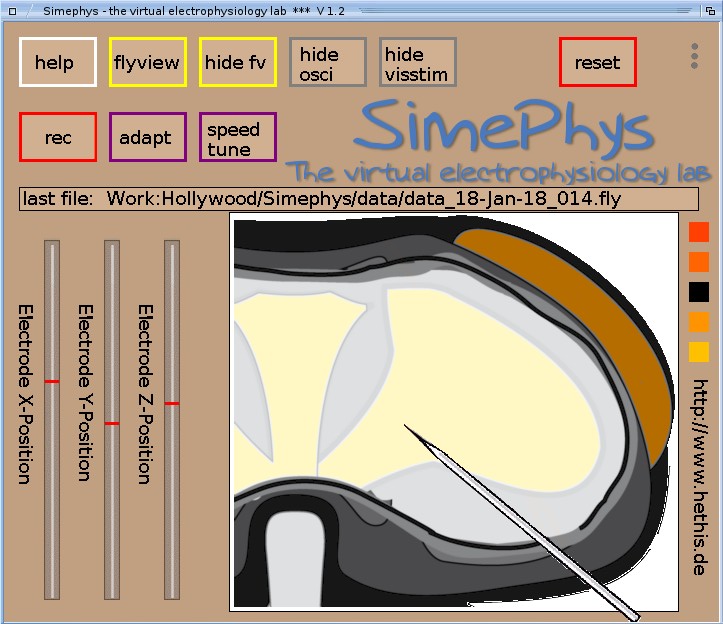
1st window: Simephys - main window, program control center and preparation
|
There are some buttons and sliders to use the program. Note, that all sliders can only be moved when the mouse pointer is over the red bar and the right mouse button is clicked while moving the slider bar. All buttons are operated by the left mouse button though. The fuctions of main window's buttons are:
At the top right of the menu (indicated by the three gray dots), there is a menu available. To open the menu, just click (left mouse button) the dots, the menu opens and vanishes after a brief time again. There are a few items available:
The function to unmask the cells requires a password. The default password is "simephys", you may change it by selecting"set password". Moving the sliders will wander the electrode along the preparation. The string below the buttons shows the name and path of the last recorded file (*.fly files). Filename consists of current calendar date and gets incremented each recording by one. If a filename does exists already the name gets incremented acoordingly until no file of identical name exists. If the full path of the filename is too long to get shown at the display the left part of the name gets cut. A cut name is indicated by two dots "..". If an update to the installed version is available a notification about that will appear just below the file name area. A mouse click on that message will open a browser window that leads to the download of the new version. |
2nd window: Visual Stimulus visual stimulus and control of it

|
With the sliders of that window the speed of the grating pattern is adjustable. The slider with the "d" is for downward speed whereas the "u" dentes the slider for the upward speed. The string field gadget the grating pattern speed additonally. |
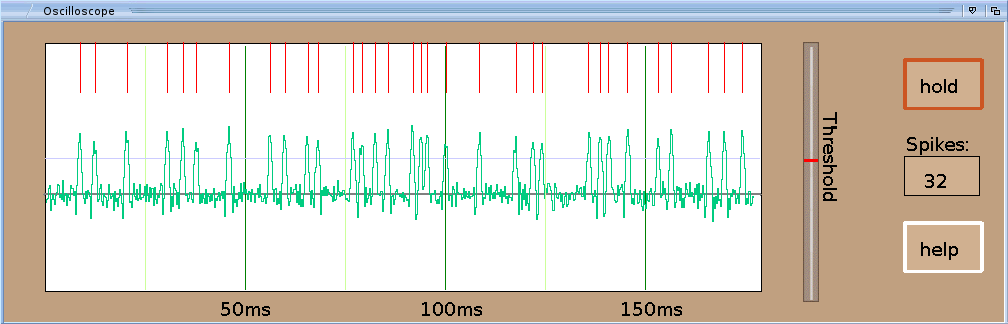
3rd window: Oscilloscope - the oscilloscope with some functions
|
The fuctions of oscilloscope window's buttons are:
The oscilloscope shows the actual cell signal recorded at the elelectrode in green. Red dashes denote detected action potentials (AP). Use the slider to apply a threshold for AP detection. The threshhold is shown as gray line at the oscilloscope screen. The string gadget shows the sum of all detected APs (spikes) within the shown 175ms time window. To start an experiment you must move the recording electrode. The electrode will move according to the coordinates of the sliders. Once you found a cell signal you can use the slider bar at the oscilloscope window to use a threshold detector to determine action potentials (APs) against background noise. The detected APs are shown as red dashes.
The button run starts a visual grating
pattern that is used for cell search and
cell type discrimination. The button turn toggles the alignment of the visual grating
(either horizontal or vertical). The adaptation named button at the main window starts an adaptation experiment. The grating pattern is moved for 0.75s with one velocity and for 0.25s with another velocity for a total of 9 times. Detected APs are recorded to harddisk for later evaluation (for example with the *.fly viewer. Data is stored in files with the .fly extension at a program's subdirectory called data. The record process is indicated by a redly tinted record button. The reset button resets the current preparation and provides another preparation and new electrode (i.e. different noise behaviour, slightly different cell positions and intrinsic properties). Don't push that button by accident, you'll lose your current preparation! To quit the application click on the quit button, the close gadget of the main window or just by press Ctrl-C. *.fly viewerThe *.fly viewer (stardotfly viewer) is a little tool to monitor and average recorded .fly data from the experiments performed with the Simephys software.The program can be run stand alone or directly from Simephys. It needs a screen resolution of at least 1024x600. If run from Simephys the binary - be named fly_view.exe or just fly_view - stay in a directory called fly_view that resides in the same directory the Simephys binary is located.
|
|
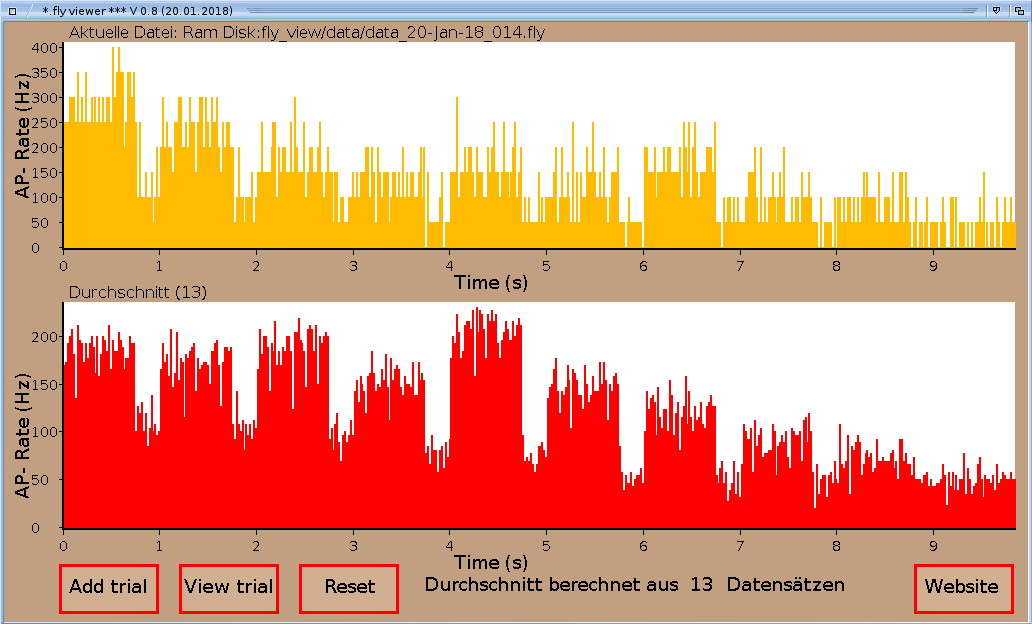
There are four buttons on bottom of the program window. The leftmost button
Add trial opens a file requester and
asks you for a .fly file.
Once a data file is chosen, the data gets displayed as a PSTH (20 ms bin width) to monitor the
action potential rate over the experiment (upper diagram). If you don't want to add a data file to your average, chose the button View trial. A file requester opens and asks you once again for a file. This time the chosen data set will not get included to the average. The current trial will be displayed in the upper trace, the average without the current trial once again in the bottom diagram. Diagrams autoscale on abscissa and ordinate to the actual data set.
The button Reset resets the current average.
Windows says this application is potentially dangerous - why that? I found a bug, want to discuss the data or methods, have a feature request or want to donate you a bag of gold ducates? Well, in that case don't hesitate to contact me.
Version 1.4.5 is a step to a largely redesigned V1.5. The main limitations of earlier versions are a fixed visual stimulus and too many fixed
logic operations within the used cell models. This should get improved! V.1.4.5 has a new stimulus system now as well as changed cell models which
use way less external controls. But this version has some major issues: Grab your copy from here:
V1.2.1 (stable)
V1.4.5 (experimental)
V1.4.5 released on 31-Dec-2022
V1.2.1 released on 28-Dec-2025
V1.2 released on 18-Jan-2018
V1.1 released on 06-Sep-2015
V1.0 released on 08-Dec-2013
V0.10 released on 29-Feb-2012
V0.9 released on 24-Jan-2011
V0.8 released on 28-Oct-2010 -- Cheers! Document history:
|
Back to main page and contact information or
to files page
--
© U. Beckers